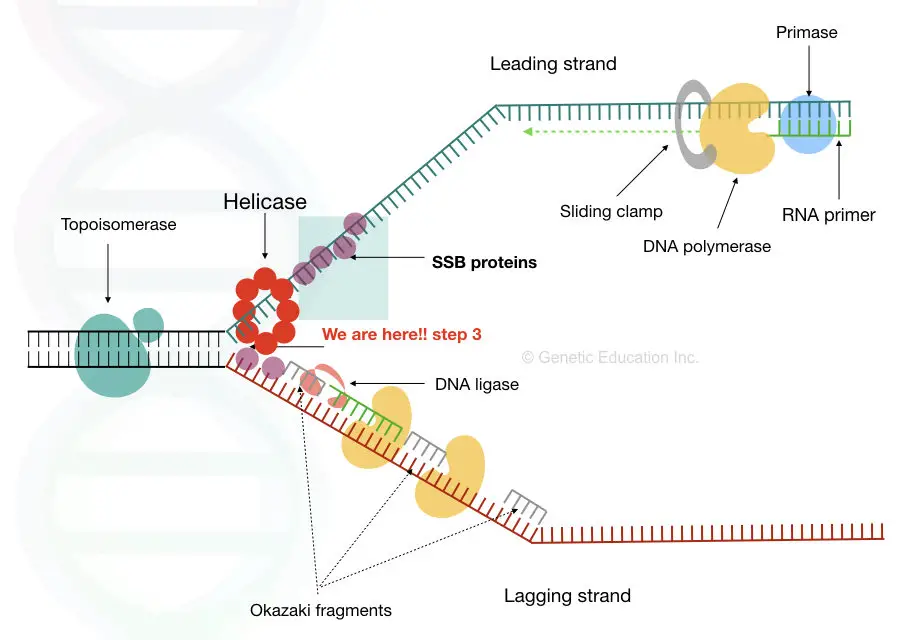
Single-stranded binding proteins ( SSBs) are a class of proteins that have been identified in both viruses and organisms from bacteria to humans. Viral SSB Although the overall picture of human cytomegalovirus (HHV-5) DNA synthesis appears typical of the herpesviruses, some novel features are emerging. Structure Single-strand binding proteins bind to the single-stranded DNA to prevent the helix from re-forming. Primase synthesizes an RNA primer. DNA polymerase III uses this primer to synthesize the daughter DNA strand. On the leading strand, DNA is synthesized continuously, whereas on the lagging strand, DNA is synthesized in short stretches called.
SingleStranded Binding Protein (SSB) Structure And Function
Single-Stranded DNA-Binding Proteins (SSBs) A.L. Eggler, in Encyclopedia of Genetics, 2001 Structure of SSBs: How Do They Bind DNA? SSBs from all organisms are, for the most part, functional homologs. Sequence similarity between various SSBs is limited; however, they share a DNA-binding motif called the OB fold. This process takes us from one starting molecule to two "daughter" molecules, with each newly formed double helix containing one new and one old strand. In a sense, that's all there is to DNA replication! But what's actually most interesting about this process is how it's carried out in a cell. In E. coli, this protein is a tetramer known as single-stranded-binding (SSB) protein, which the ssDNA winds itself around, while in T4 and T7, these proteins are monomers known as gp32 and gp2.5, respectively. Biochemical study indicates that RPA can bind ssDNA in three states (illustrated in Figure 7 (b) ): (1) binding of OB folds A and B occludes about 10 nucleotides, (2) binding of OB folds A, B and C occludes 12-23 nucleotides, and (3) binding of the four OB folds occludes 28-30 nucleotides ( Arunkumar et al., 2003; Bastin-Shanower and Brill, 2001.

Single stranded DNAbinding protein Stock Image F009/6459 Science
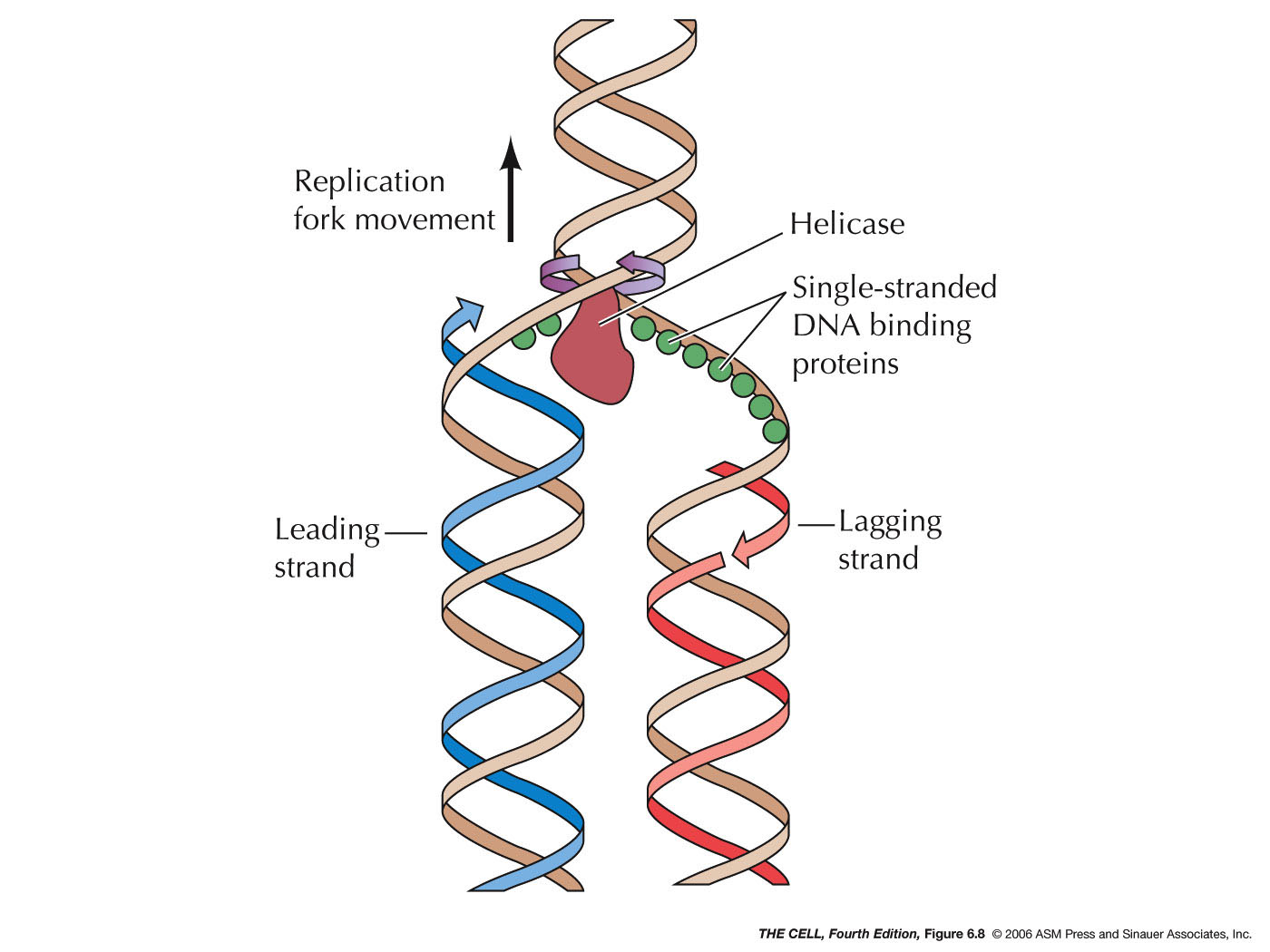
Single-strand binding proteins bind to the single-stranded DNA near the replication fork to keep the fork open. Primase synthesizes an RNA primer to initiate synthesis by DNA polymerase, which can add nucleotides only in the 5' to 3' direction. One strand is synthesized continuously in the direction of the replication fork; this is called the. Single-stranded DNA (ssDNA) binding proteins (SSBs) are critical in maintaining genome stability by protecting the transient existence of ssDNA from damage during essential biological processes, such as DNA replication and gene transcription. The single-stranded region of telomeres also requires protection by ssDNA binding proteins from being. In bacteria, these proteins are known as single-stranded DNA binding proteins (SSBs) (7, 8). Escherichia coli SSB is a homotetramer, Figure 1A. Each monomer features a structured DNA-binding domain (residues 1-112) and a long and disordered C-terminal tail (residues 116-177) containing a highly acidic tip. Single strand binding protein (ssb protein) binds to separated strands of DNA and prevents reannealing. Primase complexes with helicase, creates RNA primers (pppAC(N) 7-10) on the strands of the open duplex 2 (Primase+helicase constitute the Primosome).
DNA Replication
Single-strand DNA-binding protein ( SSB) is a protein found in Escherichia coli ( E. coli) bacteria, that binds to single-stranded regions of deoxyribonucleic acid ( DNA ). [1] Single-stranded DNA is produced during all aspects of DNA metabolism: replication, recombination, and repair. The Escherichia coli single‐strand DNA binding protein (SSB) is essential to viability where it functions to regulate SSB interactome function. Here it binds to single‐stranded DNA and to target proteins that comprise the interactome. The region of SSB that links these two essential protein functions is the intrinsically disordered linker.
Using single-molecule imaging and manipulation, the authors show linker histone H1 preferentially forms phase-separated droplets with single-stranded nucleic acids over double-stranded DNA and. The recognition of single-stranded DNA (ssDNA) is integral to myriad cellular functions. In eukaryotes, ssDNA is present stably at the ends of chromosomes and at some promoter elements. Furthermore, it is formed transiently by several cellular processes including telomere synthesis, transcription, and DNA replication, recombination, and repair.

Structural dynamics of E. coli singlestranded DNA binding protein
In one model, semiconservative replication, the two strands of the double helix separate during DNA replication, and each strand serves as a template from which the new complementary strand is copied; after replication, each double-stranded DNA includes one parental or "old" strand and one "new" strand. Single-stranded DNA (ssDNA) in a cell is highly prone to degradation, so cells shield ssDNA from nucleases by coating it with ssDNA-binding proteins (SSBs). In eukaryotes, the canonical SSB is a.




